In this blog, you will learn how to ingest changed data from PostgreSQL WAL data and store it in Amazon Simple Storage Service (Amazon S3) using a Postgres CDC pipeline in StreamSets.
Consider the use case where you have data being constantly updated in your PostgreSQL database and you want to extract the changed data in real-time and store that in Amazon S3. Using StreamSets Data Collector Engine, I will show you a very quick and easy example of a Postgres CDC pipeline to do just that. If S3 is not your preferred destination or you want to write that data to additional destinations, StreamSets makes it just as easy to write copy into Snowflake, Delta Lake on Databricks or both in a single CDC pipeline!
Postgres CDC Data Pipeline Implementation
Let’s take a look at our data pipeline implementation.
Data Pipeline
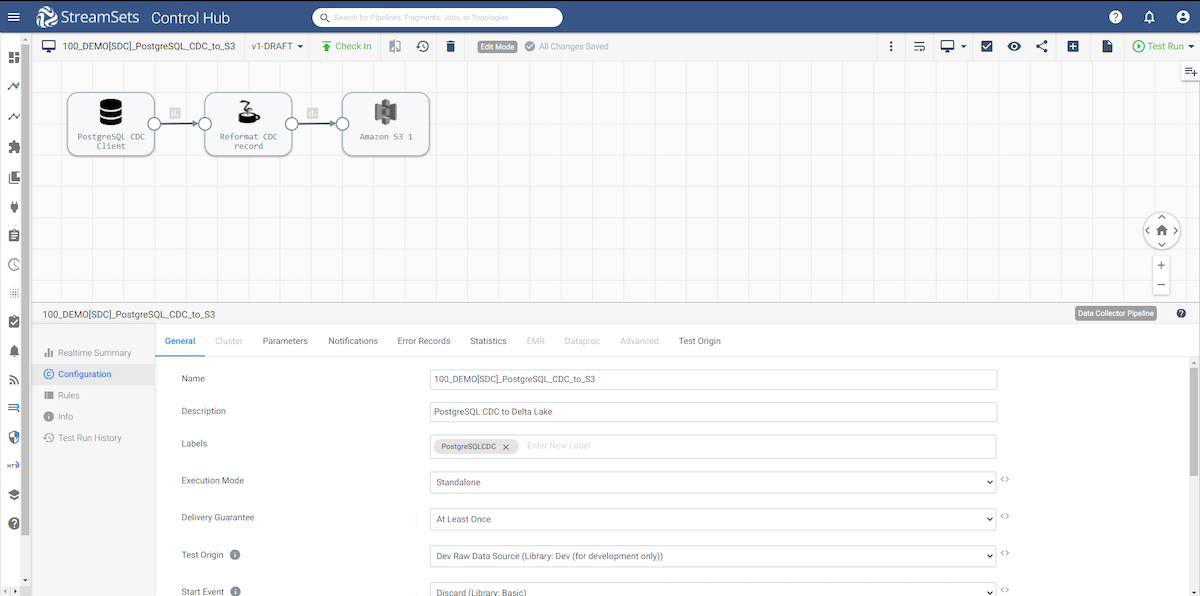
As you can see, the pipeline is very straightforward. I am using a PostgreSQL CDC Client origin, passing the data to a Jython processor to re-format the data coming from the PostgreSQL WAL log to something much easier to work with and finally writing the data to Amazon S3. Let’s take a deeper look at each stage in the pipeline.
PostgreSQL CDC Client Origin
- The Postgres CDC Client origin processes Write-Ahead Logging (WAL) data to generate change data capture records for a PostgreSQL database.
- You might use this origin to perform database replication. You can use a separate pipeline with the JDBC Query Consumer or JDBC Multitable Consumer origin to read existing data. Then start a pipeline with the PostgreSQL CDC Client origin to process subsequent changes.
- The PostgreSQL CDC Client generates a single record from each transaction. Since each transaction can include multiple CRUD operations, the PostgreSQL CDC Client origin can also include multiple operations in a record.
- When you configure the Postgres CDC Client, you configure the change capture details, such as the schema and tables to read from, the initial change to use, and the operations to include.
- You define the name for the replication slot to be used, and specify whether to remove replication slots on close. You can also specify the behavior when the origin encounters an unsupported data type and include the data for those fields in the record as unparsed strings. When the source database has high-precision timestamps, you can configure the origin to write string values rather than datetime values to maintain the precision.
- To determine how the origin connects to the database, you specify connection information, a query interval, number of retries, and any custom JDBC configuration properties that you need. You can configure advanced connection properties.
Note: The user that connects to the database must have the replication or superuser role.
PostgreSQL Prerequisite
- To enable the PostgreSQL CDC Client origin to read Write-Ahead Logging (WAL) changed data capture information, you must install the wal2json logical decoder. Install wal2json on every PostgreSQL instance being monitored for changes.
- StreamSets provides the wal2json logical decoder on GitHub. To install the wal2json, follow the instructions in the “Build and Install” section of the README.md file.
- Then, follow the configuration instructions in the “Configuration” section of the README.md file.
Let’s take a look at the Postgres CDC Client origin. (You can learn more about building data pipelines with StreamSets.)
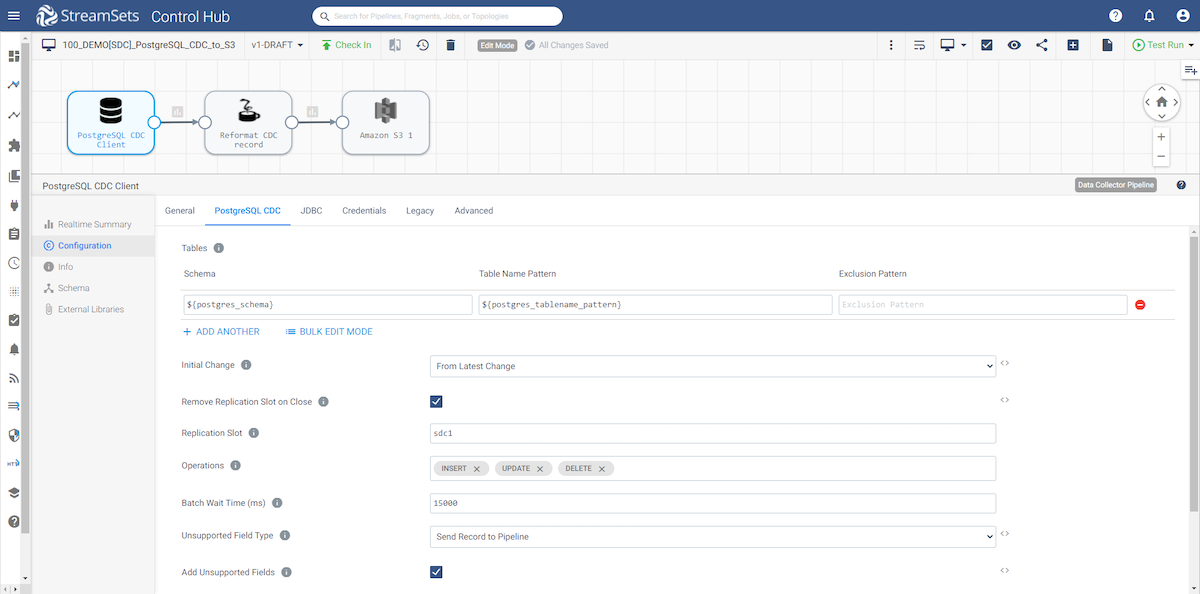
PostgreSQL CDC tab
- Here you will define the schema, a table name pattern, and an optional exclusion pattern.
- The initial change is the point in the Write-Ahead Logging (WAL) data where you want to start processing. When you start the pipeline, Postgres CDC Client starts processing from the specified initial change and continues until you stop the pipeline.
- Enter your replication slot name.
- For more information on the PostgreSQL CDC Client origin, please refer to the documentation.
JDBC tab
- Enter your JDBC Connection String using the following syntax: jdbc:postgresql://<host>:<port>/<dbname>
Credentials tab
- Enter your username and password for the PostgreSQL user with appropriate rights.
StreamSets enables data engineers to build end-to-end smart data pipelines. Spend your time building, enabling and innovating instead of maintaining, rewriting and fixing.
Jython Evaluator Processor
The Jython Evaluator processor uses custom Jython code to process data. We will use this to reformat the WAL data into something much easier to handle. For more information on the Jython Evaluator processor, please refer to the documentation.
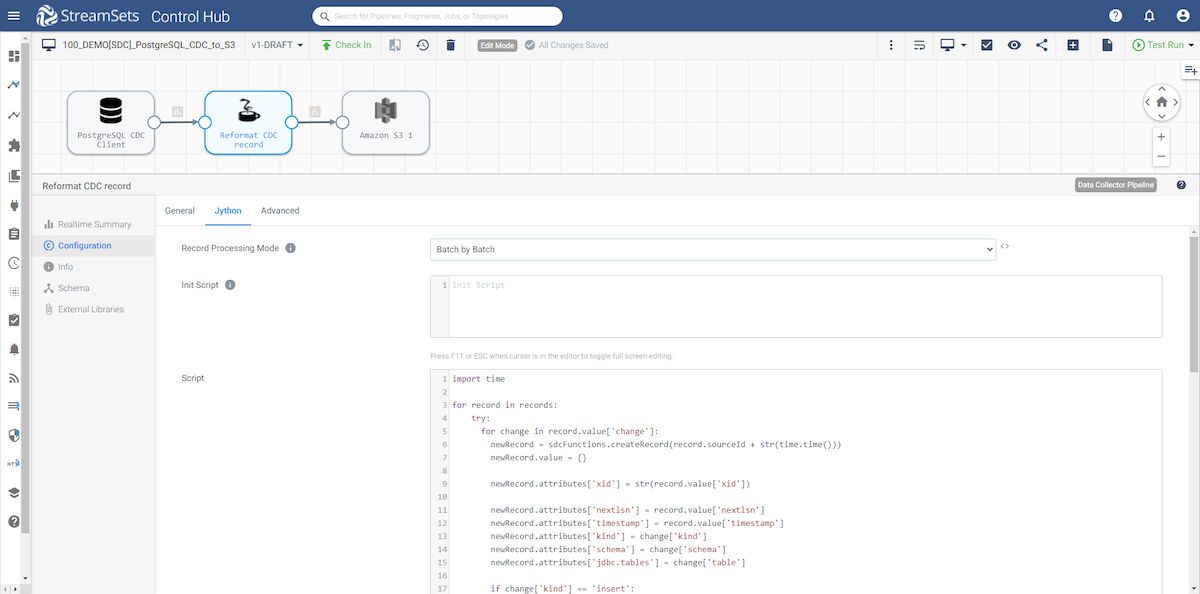
Jython tab
We are using the following Jython code to reformat the record coming from the WAL.
import time
for record in records:
try:
for change in record.value['change']:
newRecord = sdcFunctions.createRecord(record.sourceId + str(time.time()))
newRecord.value = {}
newRecord.attributes['xid'] = str(record.value['xid'])
newRecord.attributes['nextlsn'] = record.value['nextlsn']
newRecord.attributes['timestamp'] = record.value['timestamp']
newRecord.attributes['kind'] = change['kind']
newRecord.attributes['schema'] = change['schema']
newRecord.attributes['jdbc.tables'] = change['table']
if change['kind'] == 'insert':
newRecord.attributes['sdc.operation.type'] = '1'
if change['kind'] == 'delete':
newRecord.attributes['sdc.operation.type'] = '2'
if change['kind'] == 'update':
newRecord.attributes['sdc.operation.type'] = '3'
if 'columnnames' in change:
columns = change['columnnames']
types = change['columntypes']
values = change['columnvalues']
else:
columns = change['oldkeys']['keynames']
types = change['oldkeys']['keytypes']
values = change['oldkeys']['keyvalues']
for j in range(len(columns)):
name = columns[j]
type = types[j]
value = values[j]
# add data type conversions here.
if 'numeric' in type or 'float' in type or 'double precision' in type:
newRecord.value[name] = float(value) # python float == java double
else:
newRecord.value[name] = value
output.write(newRecord)
except Exception as e:
# Send record to error
error.write(record, str(e))
Amazon S3 Destination
The Amazon S3 destination is configured to store the data in JSON format. All of your S3 connection information will be configured in the Amazon S3 tab. For more information on the Amazon S3 destination, please refer to the documentation.
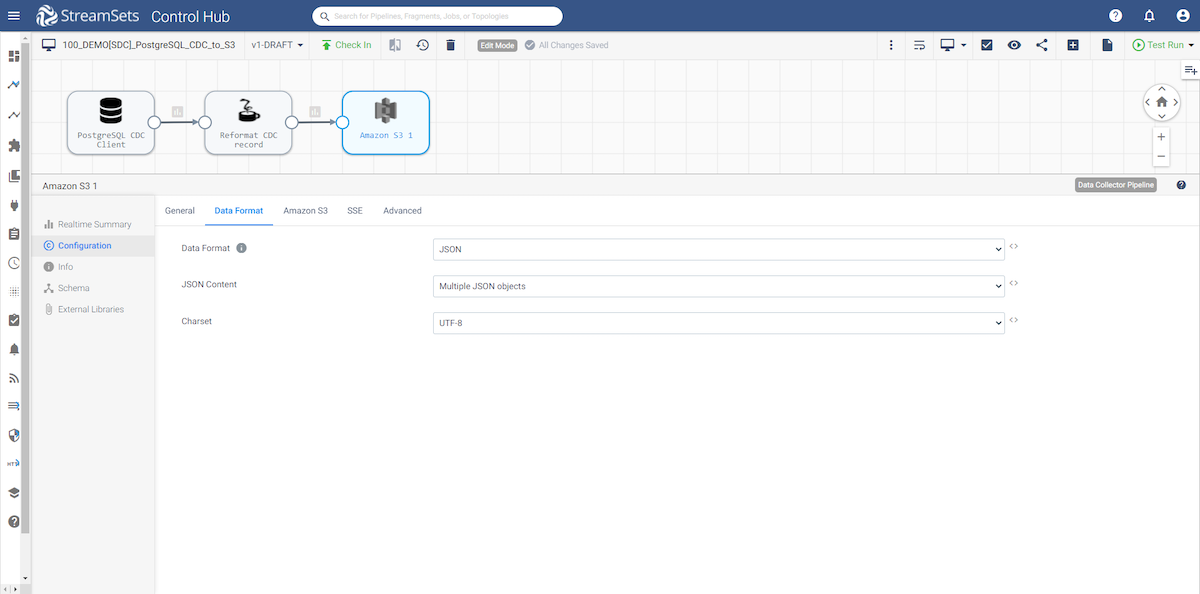
Postgres CDC Data Pipeline Run
Once you start the pipeline, make changes to the PostgreSQL table and you will see those records get processed and written to your Amazon S3 bucket.
Summary
In this post, you learned the value that can be realized by leveraging and integrating data from PostgreSQL and AWS S3 using a Postgres CDC pipeline in StreamSets. A seamless integration between PostgreSQL and AWS S3 opens up various opportunities for companies to develop new ways of accessing, analyzing, and storing their data.
Here are some resources to help jump start your journey to the cloud:
- Learn more about StreamSets for Amazon Web Services
- Getting started videos for building data pipelines
- Start building smart data pipelines today.
